The Experiment Object¶
The Experiment class provides a handy wrapper around the Consortium object with two key objectives:
- Store the state and perturbations occurred in a Consortium.
- Run sequences of dFBA + perturbations directly from configuration files.
- Automatic filtering of output files.
Using the package, we realized that a common operation was to initialize models from a directory with random initial biomasses, instantiate medium and run a loop that adds a perturbation and runs the simulation. That’s exactly the objective of this class.
Instantiating the Experiment¶
Experiment is a subclass of Consortium and follows a similar initialization, adjusted to read directly from files at the time of instantiating:
-
class
Experiment(medium_path = "", models_dir = "", rand_biomasses = [0.0001,0.0005], perturbations = [], ..., lp = "fba", solver = "glpk")¶ Parameters: - medium_path (string) – path to a JSON file containing medium and perturbations
- models_dir (string) – path to the directory where the models are read from
- rand_biomasses (list) – of two floats, upper and lower constraints to randomized initial biomasses (default = [0.0001,0.0005])
- perturbations (list) – of names of the perturbations (default = list of generic names)
- lp (string) – establishing the type of LP problem (default = “fba”) to solve by the solver (default = “glpk”)
- .. – Rest of parameters of a
Consortium.
The JSON file where medium_path points must have the structure of a list of dictionaries.
[
{
Metabolite_of_media1 : amount,
Metabolite_of_media2 : amount
...
},
{
Metabolite_of_pertubation1 : amount,
Metabolite_of_pertubation2 : amount
...
},
{
Metabolite_another_pertubation1 : amount,
Metabolite_another_pertubation2 : amount
},
...
]
The perturbations parameters must have the same length of this JSON list. Either way, a warning will be printed and, if needed, it will be filled with generic names. On the other hand, the models on models_dir directory should have an xml, mat or json extension. If a file in this directory isn’t a model, a warning will be printed.
from mmodes import Experiment
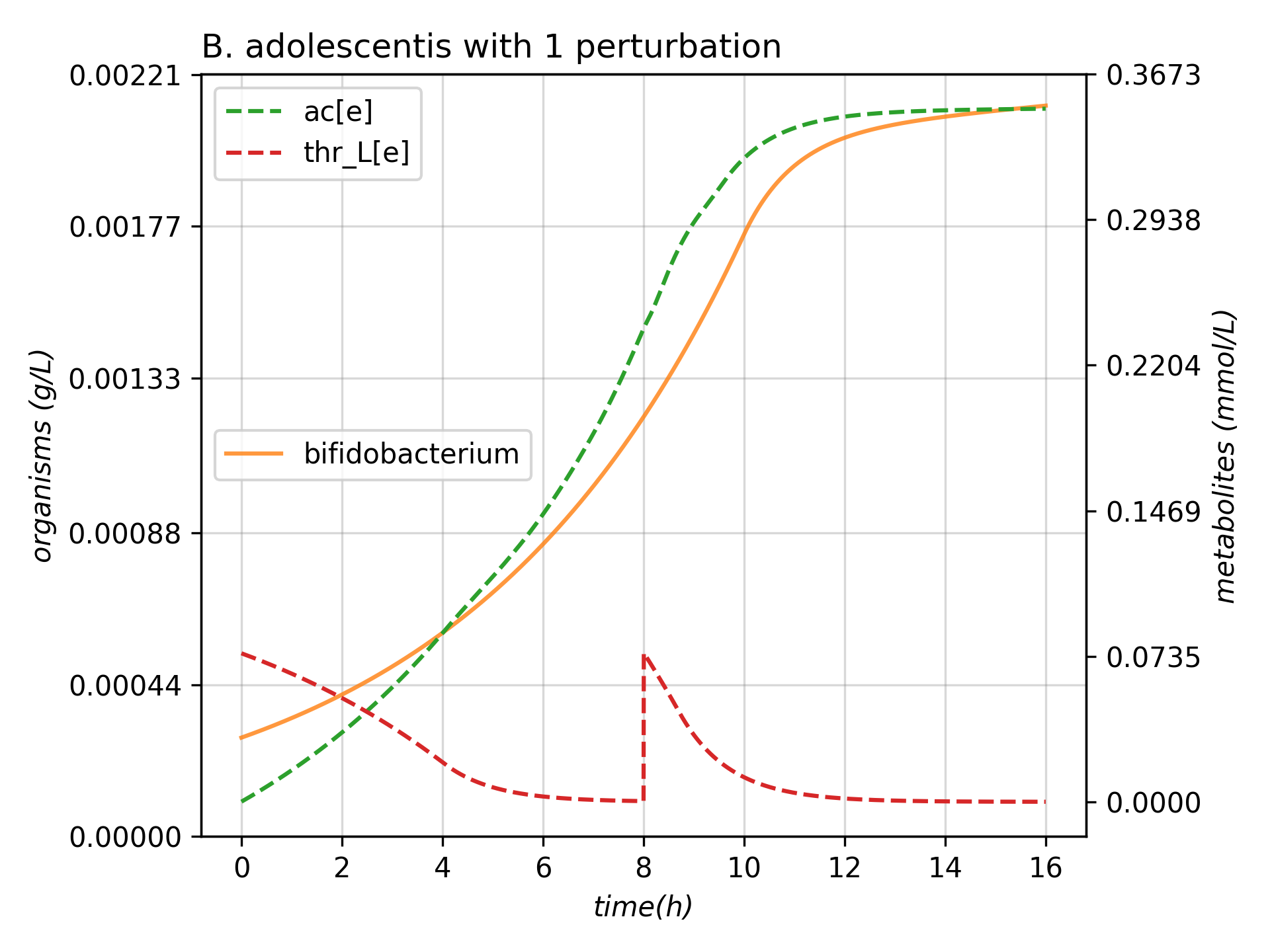
exp = Experiment(medium_path = "ModelsInput/media.json", models_dir = "ModelsInput",
perturbations = ['START', 'THR+TYR'], mets_to_plot = ["ac[e]", "thr_L[e]"],
title = "B. adolescentis with 1 perturbation")
Note
Don’t get frightened by the walls of text in the screen. Each file is tried to be read by COBRAPy with different parsers and libSBML is really verbose right now. Sadly, it can’t be filtered by now.
Running an Experiment¶
-
Experiment.run_experiment(intervl = 10, integrator = 'vode', stepChoiceLevel = (), verbose = False, outp = "models_dir.png", filter = False, equif = True, inplace_filter = False, plot = True, actualize_every = float(-inf))¶ Starts a loop of community simulations + perturbations
Parameters: - intervl (float) – time in simulation units (hours) between perturbations (default = ‘10’).
- integrator (str) – (‘vode’ ‘dopri5’ ‘fea’ ‘rk4’) type of ODE integrator (default = ‘vode’).
- stepChoiceLevel (str) – (0, max time step, max number of time-steps) for vode and (time-step, 0, max number of time-steps) for the rest of integrators (default = 0., 0.5, 100).
- verbose (bool) – a verbose simulation will show a progress bar, the reason of exiting the simulation and some messages of perturbations (default = False).
- out (string) – path where the output will be generated (default= plot.tsv).
- outp (string) – path where the plot will be generated (default= “Some_Experiment.png”).
- plot (bool) – whether to generate the plot (default= True).
- actualize_every (float) – time interval of writing to output files (default = -inf, write always)
The rest of parameters are discussed in the next section.
#16 h of simulation
exp.run_experiment(intervl = 8, integrator = 'fea', stepChoiceLevel = (0.005,0.5,10000))
It should have generated a tsv called plot.tsv and an image called Some_Experiment.png like the one presented:
Filtering the output¶
The 3rd objective of this class was to filter the output. But how exactly is filetered? Briefly, the points right before each Perturbation in Medium output and fluxes are kept. In addition, the fluxes can be filtered by keeping 100 equidistant points, generating other file. We found that those kind of files were really useful for some applications.
run_experiment().| param bool filter: | |
|---|---|
| whether output should be filtered (default = False) | |
| param bool equif: | |
| whether the equidistant flux output should be generated. It only works when filter is True (default = True). | |
| param bool inplace_filter: | |
| whether the original output should be overwritten by the filtered one (default = False) | |