Getting started¶
An example script is provided on the GitHub repository and will be here described here.
First steps: the Consortium object¶
You need a GEM model to run a consortium dynamic simulation. BiGG models is a good place to start. Conversely, a model example of Bifidobacterium adolescentis of the AGORA database is provided as example on the ModelsInput.
from mmodes import Consortium, dMetabolite
cons = Consortium(mets_to_plot = ["thr_L[e]", "ac[e]"])
mets_to_plot parameter is supplied to later plot these metabolites. Now, we add the model to the Consortium object.
path_to_model = 'mmodes/ModelsInput/BGN4_eu.xml' # provided example GEM
# Additionally, we'll instantiate some metabolite
# to assign kinetic parameters
# for instance, https://www.ncbi.nlm.nih.gov/pubmed/18791026?dopt=Abstract
glc = dMetabolite(id = "glc_D[e]", Km = 14.8, Vmax = 0.13)
cons.add_model(path_to_model, 0.0003, dMets = {glc.id: glc})
More information of Consortium class can be found on The Consortium Object.
Instantiating the medium¶
The last step previous to running the simulation is assigning a medium to the Consortium. In the example, medium is read from a JSON file (which is a dictionary).
abs_media = {k: 1000 for k in cons.media}
cons.media = cons.set_media(abs_media)
print(cons)
Make sure that all metabolites in the medium are named the same as in the added GEM models. MMODES supports working with metabolite identifiers or names, default is id. If some metabolites are misspelled, they won’t be added. Conversely, metabolites that are in the extracellular compartment of the GEM members of the Consortium, that were not added to the medium, will be set to 0.
Run the simulation!¶
The last step is running the simulation.
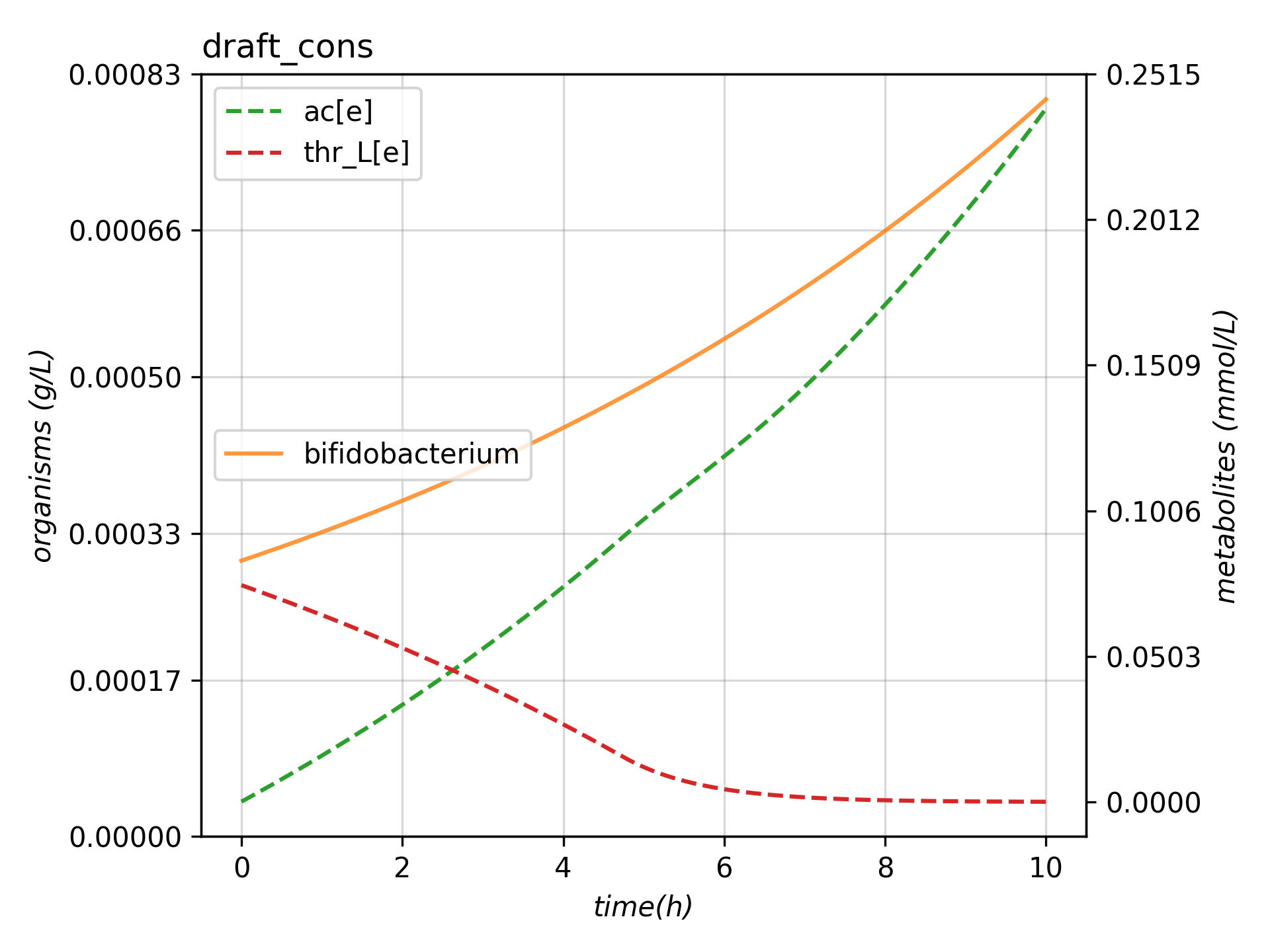
cons.run(maxT = 10, outp = "plot_example.png", outf = "tab_example.tsv", verbose=True)
# print information pieces on screen
for mod in cons.models:
print(mod, cons.models[mod].volume.q, sep = " -> ")
print("Glucose", str(cons.media["glc_D[e]"]), sep = " -> ")
print("Acetate", str(cons.media["ac[e]"]), sep = " -> ", end = "\n\n")
As demonstrated, MMODES allows running a dynamic (p)FBA simulation with a few lines of code. A TSV file and a plot should’ve been generated on the working directory.
Also, The Experiment Object demonstrates how the Consortium class can be extended to ease even more the configuration of microbial community simulations.